User login
With an increased incidence of 13.1 per 1000 inpatients1, 2 and an attributable mortality of 6.1%,3 in 2006 the Canadian government added Clostridium difficileassociated disease (CDAD) to its list of reportable diseases.4 The Centers for Disease Control and Prevention and the Infectious Disease Society of America subsequently created definitions of the disease and recommended surveillance of rates of health care facilityassociated CDAD, which has been found to double a patient's length of stay and cost of hospitalization.57 While not included in the current list of hospital‐acquired conditions for which payment is declined,8 the Centers for Medicare & Medicaid Services (CMS) has noted CDAD as under consideration for addition to the list,9 which would require hospital reporting of CDAD rates. This reporting is typically a labor‐intensive, medical record review process that increases the cost of delivering health care. Health care institutions have reported spending as much as $21 million per year or $400 per discharged patient on quality improvement.10 These costs are passed on to payers such as those governed by the CMS that are projected to pay for more than half of all national health spending in 2018.11 Therefore, it is prudent to examine alternatives for determining rates of disease for public reporting and quality improvement initiatives such as infection control and antibiotic stewardship programs.
There are 3 methods that can be used for this reporting. First, medical record review has been used for determining the case rate of CDAD by published reports.1, 2 This procedure allows the CMS to define the desired data to be collected, but it is labor‐intensive. Second, the use of International Statistical Classification of Diseases and Related Health Problems, 9th Edition (ICD‐9) codes from hospital databases offers the speed of database query. However, due to diagnostic and coding errors, it may report inaccurate rates of disease. Previous reports include a university‐based hospital that found a sensitivity of 78% and specificity of 99.7%,12 whereas a Veterans Administration medical center found nearly two‐thirds of patients with CDAD did not have the ICD‐9 code for C. difficile infection noted in their database.13 Therefore, the accuracy of ICD‐9 code at a community hospital that better represents the majority of United States hospitals is needed. The third method of identifying CDAD is the presence of toxin identified in the microbiology laboratory. Although this method offers ease of obtainment, it suffers from potential inaccuracy arising from duplicate patient samples, positive samples in patients who are symptom‐free carriers, and patients with CDAD despite having a negative toxin.
The same organizations that recommend surveillance for CDAD have identified that there are inadequate data on which to base a decision regarding how to proceed with routine community and hospital surveillance.6 In the setting of public reporting and nonpayment, the method of identifying CDAD cases must be accurate to ensure fairness, while being inexpensive. To identify the potential value of these less labor‐intensive methods of reporting the incidence of CDAD, we evaluated the use of ICD‐9 codes and C. difficile toxin to accurately report the incidence of CDAD at a community hospital, as well as the labor hours required for each reporting method.
METHODS
Patients >18 years of age and potentially having CDAD were identified via database queries for ICD‐9 codes and positive C. difficile toxin assays at our institution from November 1, 2006, through August 31, 2007. Our institution is a 379‐bed university‐affiliated community teaching hospital in a socio‐economically diverse area of Baltimorewith approximately 110,000 emergency department visits, 30,000 discharges, and 110,000 inpatient days each yearthat uses a handwritten paper chart for all provider orders and patient documentation. The C. difficile toxin assay method used during the study period was an enzyme immunoassay that detects both toxins A and B (Meridan Bioscience, Cincinnati, OH). ICD‐9 codes were queried in the CareScience database(Premier, Inc; Charlotte, NC), while C. difficile toxin was queried via the hospital laboratory database. All identified patients underwent a medical record review to confirm the diagnosis of CDAD and the patient's location at the time of disease acquisition. To eliminate duplicate reporting of a single episode of disease, all duplicate patient visits with a diagnosis of CDAD within 30 days were removed.
Our diagnostic criteria (Table 1) used the recommended criteria of a combination of symptoms and positive toxin assay, visualization of pseudomembranes on colonoscopy or pathology‐proven CDAD.6 Based on the presence of CDAD in 25% of patients with a white blood cell count >30,000 and the recommendation that CDAD be considered in all patients with a white blood cell count >15,000, we included the criterion of clinical findings with severe leukocytosis to identify patients with toxin‐negative CDAD.14
| Chart Review Criteria for CDAD | Chart Review Criteria for CDAD Obtained While Patient Was Not at Our Hospital | Chart Review Criteria for CDAD Obtained While Patient Was at Our Hospital |
|---|---|---|
| ||
| Pseudomembranous colitis seen during endoscopy OR biopsy or resection with surgical pathology consistent with CDAD | One of the above symptoms or objective criteria positive within the first 3 days of hospitalization AND patient was not cared for at our insitution within the last 7 days | Patient was cared for at our institution within 7 days of presentation OR the patient's symptoms were not present upon arrival at the hospital and the symptoms began after the third day of hospitalization |
| If neither of the above are present, then Clostridium difficile toxin or WBC count >25,000 plus one of the following: | ||
| Diarrhea | ||
| Fever without other cause | ||
| Abdominal pain without other cause | ||
| Colonic ileus without other cause | ||
Patients were identified by querying the CareScience database for patients having an ICD‐9 code of 008.45. Duplicate cases for the same patient within 30 days were removed. These cases were compared with the cases of CDAD determined by the medical record review for analysis. Patients were identified via positive C. difficile toxin by querying the microbiology laboratory database. A query of all patients with a positive C. difficile toxin was performed. Duplicate samples for the same patient within 30 days were removed. Patients are considered to be outside the hospital when CDAD was acquired if their toxin was positive within the first 3 days of arrival to the hospital, and they were not hospitalized at our institution within 1 week before arrival. All other cases were considered to be acquired while in our hospital. The number of patients with a toxin‐positive stool sample was compared with the medical record review.
Recognizing that it is unrealistic to review the medical records of the 23,495 discharges during the 10‐month study period, a random sample review of 500 charts not included in our CDAD‐identified patient list was performed to identify the rate at which CDAD patients failed to be identified by our methods. From a list of discharges during the study period, as long as they were not in our study population, every thirtieth patient was selected for review up to a total of 500 patient charts. Then, the identified case rate was used to predict the prevalence of disease at our institution during the study period.
Sample selection was based on the absence of previous studies evaluating the performance of C. difficile toxin assay, and the desire to have the same assay used during the entire study. The lack of data from which to perform sample size determination would result in an inaccurate estimate. Therefore, we chose a period that offered the maximum sample size, during which a single assay was used at our institution. Compared with medical record review, the accuracy and positive predictive values of the ICD‐9 code and positive stool toxin methods to identify the cases of CDAD were compared. A true positive was when the ICD‐9 or laboratory query method identified a patient who had CDAD based on chart review; a false positive was when the ICD‐9 or laboratory query method identified a patient who did not have CDAD based on chart review. Rates of over‐ or underdiagnosis, case rates, and acquisition location were determined. Statistical analysis of the case rates and acquisition location were performed via chi‐square test. The time to perform these queries was collected with accuracy to the minute.
RESULTS
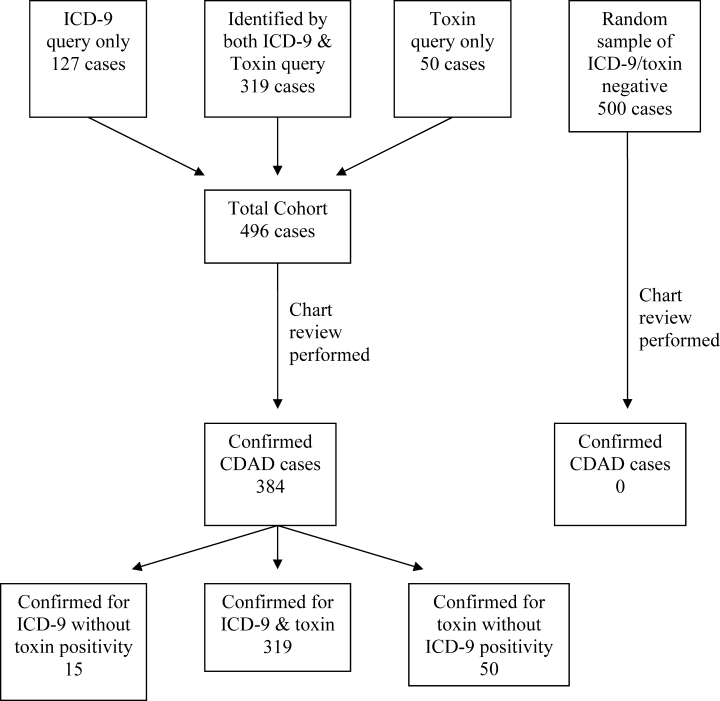
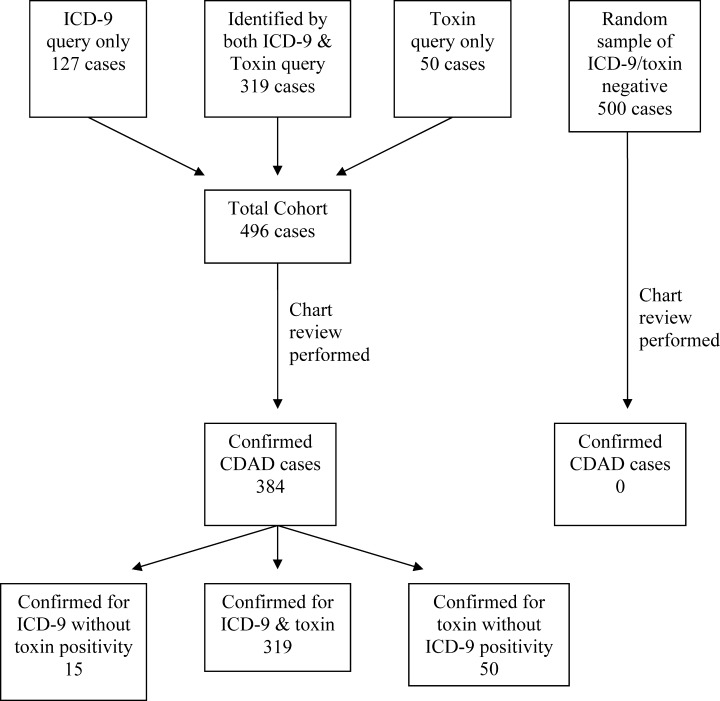
Of the 23,495 discharges during the study period, the combination of ICD‐9 and C. difficile toxin assay identified a total cohort of 496 patients, 319 of whom were identified by both the ICD‐9 method and the toxin assay, 50 of whom were identified only by the toxin assay, and 127 of whom were identified only by the ICD‐9 method. Chart review confirmed the presence of CDAD in 384 of these 496 cases, for a case rate of 16.3 per 1000 discharged patients (Table 2). The diagnostic criteria for each of these confirmed cases are listed in Table 3. Of the 384 confirmed CDAD cases, 319 were identified by both the ICD‐9 and toxin assay, 50 were identified only by the toxin assay, and 15 were identified only by the ICD‐9 query. Of the 50 cases identified by the toxin assay that were not identified via the ICD‐9 method, all 50 (100%) were confirmed to have CDAD by chart review. In contrast, of the 127 cases identified via the ICD‐9 method that were not found via the toxin assay, only 15 (11.8%) were confirmed to have CDAD by chart review (Figure 1).
| ICD‐9 | Toxin Assay | Chart Review | |
|---|---|---|---|
| |||
| No. of patients identified | 446 | 369 | 384 |
| Case rate per 1000 discharges* | 19.0 | 15.7 | 16.3 |
| 95% confidence interval | 17.320.8 | 14.117.4 | NA |
| Case rate compared with chart review, P | P = 0.001 | P = 0.440 | NA |
| CDAD rate reported compared with chart review | 116.1% | 96.1% | NA |
| Accuracy | 83.9% | 96.1% | NA |
| PPV | 74.9% | 100% | NA |
| Portion of cases acquired at our hospital, % | NA | 48.2% | 40.6% |
| Portion of cases acquired at our hospital, P | NA | P = 0.003 | NA |
| Minutes consumed for data collection | 312 | 842 | 21,899 |
| Estimated annual cost per hospital | $234.00 | $631.50 | $16,424.25 |
| Criterion | No. of Cases | Case Rate* |
|---|---|---|
| ||
| Endoscopy | 2 | 0.5% |
| Surgical pathology | 9 | 2.3% |
| Positive toxin and diarrhea | 369 | 96.1% |
| WBC >25.0 and diarrhea | 51 | 13.3% |
| WBC >25.0 and fever without other source | 9 | 2.3% |
| WBC >25.0 and unexplained abdominal pain | 17 | 4.4% |
| WBC >25.0 and colonic ileus | 2 | 0.5% |
Of these 384 cases, 369 were identified via the toxin assay for a case rate of 15.7 per 1000 patient discharges, which was not found to be different from the rate of 16.3 determined by chart review (P = 0.440; 95% confidence interval [CI], 14.117.4). Compared with chart review, the toxin assay reported 96.1% (369/384) of cases. Chart review demonstrated that every patient who had a positive toxin assay met the diagnostic criteria for CDAD for a positive predictive value (PPV) of 100%.
The ICD‐9 method identified 446 patients thought to have CDAD, 334 of whom were confirmed by chart review for a PPV of 74.9% (334/446). Compared with chart review, the ICD‐9 method reported 116.1% (446/384) of cases for a case rate of 19.0 per 1000 discharges and was significantly different than the rate of 16.3 reported by chart review (P = 0.001; 95% CI, 17.320.8).
Chart review identified 156 of 384 (40.6%) patients who acquired CDAD while at our hospital and 228 of 384 (59.4%) who acquired it elsewhere. In comparison, the toxin assay criteria identified 369 cases of CDAD, of which 48.2% (178/369) were acquired while at our hospital and 51.8% (191/369) were acquired elsewhere (P = 0.003).
The time for data extraction via these 3 methods differed greatly. The ICD‐9 method only consumed 312 minutes and the toxin assay method 842 minutes, whereas the chart review method consumed 21,899 minutes. These times reported include the database query and data analysis for the ICD‐9 and toxin assay methods, while it includes the database query and list generation along with the manual chart review and data analysis for the chart review method. The review of the random sample of patients believed not to have CDAD was not included in any of the reported times. Chart review on a random sample of 500 patients not previously identified for review found no additional cases of CDAD.
DISCUSSION
Our study demonstrates that use of positive C. difficile toxin assay data from the microbiology laboratory alone is an efficient method of identifying patients with CDAD. This method only consumed 842 minutes versus 21,899 minutes consumed by the chart review method. The C. difficile toxin method reduces the workforce required to collect and analyze this data, but more importantly, it was found to be reliable by reporting an institutional case rate that is similar to that of chart review.
In contrast, the ICD‐9 method was efficient but less reliable. It only consumed 312 minutes, but it overreported the institutional rate of CDAD by 16.1%, had a PPV of only 74.9%, and of those patients who were identified by ICD‐9 but not toxin assay, only 11.8% actually had CDAD. This finding is in conflict with the previously noted underreporting of this method.13, 15 We believe this difference to be associated with institutional differences, because previous reports originated from a veterans hospital and an academic medical center, and previous authors have failed to use predefined diagnostic criteria and a complete chart review to confirm cases of CDAD. Similar to our study, an academic medical center identified that listing of CDAD in a patient's medical history in their chart was associated with a false‐positive ICD‐9 code for CDAD.15 This observation appears to bring clarity to one of the causes of the variance of ICD‐9 code accuracy between institutions. Institutions seem to vary on the method of attaching diagnoses to the patient's final hospital record. Some institutions include only what is listed as an active problem, whereas others list diagnoses listed in the chart as previous problems and those listed as a potential diagnosis without confirmation. Another potential cause of the ICD‐9 inaccuracy is the potential of clinicians to diagnose and treat a patient for CDAD in the absence of the diagnostic criteria used for chart review. Physician practices such as these are known to vary between institutions leading to a variance in the ICD‐9 code accuracy.
In total, 15 cases of CDAD were identified in the absence of a positive toxin assay, and of these, 12 cases were identified using leukocytosis‐based criteria (Table 4). This resulted in 3.1% of our cases being toxin‐negative, based on leukocytosis criteria, and is lower than the previously identified 35% of cases.14 Because it was the only assay available at the time, this previous research used a toxin Aonly assay, which is more likely to have false‐negative results than the toxin A/B assay used at our institution during the study period. The investigators also required all toxin‐negative patients to have been recently treated with antibiotics. Based on the increasing rates of community‐acquired CDAD, including those that are antibiotic‐nave patients, we felt a history of antibiotic exposure was no longer a prerequisite for CDAD and thus excluded it from our diagnostic criteria.16, 17 Based on these differences, we feel our results are likely an accurate reflection of the number of cases identified by ICD‐9 query in the absence of toxin positivity. However, concerns should be further alleviated through the realization that nonuse of this strategy improves the accuracy of the toxin assay method, while reducing the accuracy of the ICD‐9 method and thereby strengthening the validity of our conclusions. Mathematically, this would result in 369 of 369 patients identified by toxin assay and 369 patients identified by the ICD‐9 method. This would reduce the case rate of the chart review method to the same 15.7 of the toxin assay, while the ICD‐9 method would remain at 19.0.
| Criterion | No. of Cases* |
|---|---|
| |
| Endoscopy | 0 |
| Surgical pathology | 3 |
| WBC >25.0 and diarrhea | 10 |
| WBC >25.0 and fever without other source | 1 |
| WBC >25.0 and unexplained abdominal pain | 3 |
| WBC >25.0 and colonic ileus | 1 |
We considered whether the toxin assay method may overestimate the number of cases due to a C. difficileasymptomatic carrier rate as high as 50% of hospitalized patients.6 However, we found no difference in the case rate when compared with that of chart review, and there were no false‐positive cases. We believe this is attributable to the 30‐day window that was used to identify a single episode of CDAD and the absence of toxin assays being performed on asymptomatic individuals. To avoid overrepresentation of the actual number of CDAD cases, we chose to label all repeat positive toxins and repeat hospitalizations within 30 days as a single episode of CDAD. This was based on the identification that 56% of patients remained toxin‐positive 26 weeks after adequate treatment for CDAD.18 The enzyme‐linked immunosorbent assay (ELISA) method used by our laboratory is the method used to report 94% of CDAD cases in a national point prevalence study that collected data from United States acute care hospitals with representation from 47 states1, 6, 19 (Table 5). This use of ELISA in the majority of United States hospitals suggests that our data can be extrapolated for use throughout the United States. While laboratories are increasing their use of 2‐step algorithms involving glutamate dehydrogenase antigen assay followed by cytotoxin neutralization, and more recently beginning the use of polymerase chain reaction assays, both of these methods have been found to increase the accuracy of detecting C. difficile compared with ELISA.20 Therefore, as laboratories evolve to use more accurate assays to detect CDAD, the methods described herein will be expected to increase in reliability.
| Method | Sensitivity | Specificity | Cost | Ease of Performance | Typical Results Reporting | Notes |
|---|---|---|---|---|---|---|
| ||||||
| Culture | Gold standard | NA | $$$$ | Difficult | Days | Slow turnaround time; is the standard upon which other test methods are based, but not all organisms are toxin‐producing |
| Cell cytotoxicity assay | 67%100% | Gold standard | $$$ | Intermediate | Next day | Is the standard upon which other test methods are based to identify toxin‐producing stains of Clostridium difficile |
| EIA for toxin A/B | 63%94% | 75%100% | $ | Easy | Same day | Used by >90% of laboratories in the United States |
| EIA for detection of Clostridium difficile common antigen (GDH) | 85%95% | 89%99% | $ | Easy | Same day | Provides no information regarding the toxigenicity of the isolate, typically used in combination with cell cytotoxicity assay to identify toxin‐producing strains |
| Polymerase chain reaction | 96%100% | 88%91% | $$ | Intermediate | Same day | More data are needed before recommendation for routine testing |
The toxin assay methodology used to determine the rate of CDAD cases acquired while the patient was at our hospital overreported these cases. Based on this result, identification of individual cases of CDAD that are obtained at a specific hospital would continue to require manual chart review. This expensive method may be avoided by instead choosing to use institutional case rates for reporting, monitoring, and incentivizing hospitals. However, a discussion of the methods of this approach and its confluence with our societal goal to move toward Accountable Care Organizations is beyond the scope of this discussion section.
Although it appears that we identified all cases of CDAD occurring at our institution, a limitation of this study is its inability to review all charts during the 10‐month study period. We used a combination of ICD‐9 and positive C. difficile toxin assay data to identify all possible cases of C. difficile. The current approach to case identification for reported hospital conditions is limited to an ICD‐9 database query. This query is followed by chart review to collect data for hospital performance that is published in locations such as
Increased automation is expected in the future of reporting. The Centers for Disease Control and Prevention found increased rates of disease reporting and increased accuracy when reporting is electronically automated via their software system, Electronic Support for Public Health, which is designed to communicate with and perform automated data queries on providers' electronic medical records.21 While use of this model is creeping into the health system for reporting to public health authorities,22 universal hospital electronic medical record implementation and full connectivity with such reporting systems is many years from fruition. In addition to its practical use for reporting CDAD in our current health system, our work easily transitions into automated reporting within an electronically integrated health system once achieved.
In conclusion, ICD‐9 data were found to be unreliable, and consideration must be given to cessation of their use for CDAD case rate research and reporting. Use of a positive C. difficile toxin assay accurately reports the institutional incidence of disease, can be used by individual institutions to self‐monitor case rates or by the government to determine regionally acceptable intuitional rates of CDAD on which incentives and penalties can be based, and will increase in efficiency as reporting continues to be automated. This process can be instituted at a fraction of the cost of the standard chart review that is currently used for most reporting.
- ,,,.National point prevalence of Clostridium difficile in US health care facility inpatients, 2008.Am J Infect Control.2009;37:263–270.
- ,,.Secular trends in hospital‐acquired Clostridium difficile disease in the United States, 1987–2001.J Infect Dis.2004;189:1585–1589.
- ,,, et al.Analysis of 30‐day mortality for Clostridium difficile‐associated disease in the ICU setting.Chest.2007;132:418–424.
- .Final report and recommendations from the National Notifiable Diseases Working Group.Can Commun Dis Rep.2006;32:211–225.
- ,,, et al.Recommendations for surveillance of Clostridium difficile‐associated disease.Infect Control Hosp Epidemiol.2007;28:140–145.
- ,,, et al.Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA).Infect Control Hosp Epidemiol.2010;31:431–455.
- ,.Review of current literature on the economic burden of Clostridium difficile infection.Infect Control Hosp Epidemiol.2009;30:57–66.
- Centers for Medicare 35:544–550.
- ,,, et al.Health spending projections through 2018: recession effects add uncertainty to the outlook.Health Aff (Millwood).2009;28:w346–w357.
- ,,,.ICD‐9 codes and surveillance for Clostridium difficile‐associated disease.Emerg Infect Dis.2006;12:1576–1579.
- ,,,.Fluoroquinolone use and risk factors for Clostridium difficile‐associated disease within a Veterans Administration health care system.Clin Infect Dis.2007;45:1141–1151.
- ,,.Conditions associated with leukocytosis in a tertiary care hospital, with particular attention to the role of infection caused by clostridium difficile.Clin Infect Dis.2002;34:1585–1592.
- ,,,.Accuracy of ICD‐9 coding for Clostridium difficile infections: a retrospective cohort.Epidemiol Infect.2007;135:1010–1013.
- ,,,.Implications of the changing face of Clostridium difficile disease for health care practitioners.Am J Infect Control.2007;35:237–253.
- .Clostridium difficile is no longer just a nosocomial infection or an infection of adults.Int J Antimicrob Agents.2009;33(suppl 1):S42–S45.
- ,,,,.Treatment of antibiotic‐associated Clostridium difficile colitis with oral vancomycin: comparison of two dosage regimens.Am J Med.1989;86:15–19.
- ,,, et al.Comparison of real‐time PCR techniques to cytotoxigenic culture methods for diagnosing Clostridium difficile infection.J Clin Microbiol.2011;49:227–231.
- ,,,.Comparison of BD GeneOhm Cdiff real‐time PCR assay with a two‐step algorithm and a toxin A/B enzyme‐linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection.J Clin Microbiol.2010;48:109–114.
- Centers for Disease Control and Prevention.Automated detection and reporting of notifiable diseases using electronic medical records versus passive surveillance—Massachusetts, June 2006‐July 2007.MMWR Morb Mortal Wkly Rep.2008;57:373–376.
- ,,, et al.Development of an electronic public health case report using HL7 v2.5 to meet public health needs.J Am Med Inform Assoc.2010;17:34–41.
With an increased incidence of 13.1 per 1000 inpatients1, 2 and an attributable mortality of 6.1%,3 in 2006 the Canadian government added Clostridium difficileassociated disease (CDAD) to its list of reportable diseases.4 The Centers for Disease Control and Prevention and the Infectious Disease Society of America subsequently created definitions of the disease and recommended surveillance of rates of health care facilityassociated CDAD, which has been found to double a patient's length of stay and cost of hospitalization.57 While not included in the current list of hospital‐acquired conditions for which payment is declined,8 the Centers for Medicare & Medicaid Services (CMS) has noted CDAD as under consideration for addition to the list,9 which would require hospital reporting of CDAD rates. This reporting is typically a labor‐intensive, medical record review process that increases the cost of delivering health care. Health care institutions have reported spending as much as $21 million per year or $400 per discharged patient on quality improvement.10 These costs are passed on to payers such as those governed by the CMS that are projected to pay for more than half of all national health spending in 2018.11 Therefore, it is prudent to examine alternatives for determining rates of disease for public reporting and quality improvement initiatives such as infection control and antibiotic stewardship programs.
There are 3 methods that can be used for this reporting. First, medical record review has been used for determining the case rate of CDAD by published reports.1, 2 This procedure allows the CMS to define the desired data to be collected, but it is labor‐intensive. Second, the use of International Statistical Classification of Diseases and Related Health Problems, 9th Edition (ICD‐9) codes from hospital databases offers the speed of database query. However, due to diagnostic and coding errors, it may report inaccurate rates of disease. Previous reports include a university‐based hospital that found a sensitivity of 78% and specificity of 99.7%,12 whereas a Veterans Administration medical center found nearly two‐thirds of patients with CDAD did not have the ICD‐9 code for C. difficile infection noted in their database.13 Therefore, the accuracy of ICD‐9 code at a community hospital that better represents the majority of United States hospitals is needed. The third method of identifying CDAD is the presence of toxin identified in the microbiology laboratory. Although this method offers ease of obtainment, it suffers from potential inaccuracy arising from duplicate patient samples, positive samples in patients who are symptom‐free carriers, and patients with CDAD despite having a negative toxin.
The same organizations that recommend surveillance for CDAD have identified that there are inadequate data on which to base a decision regarding how to proceed with routine community and hospital surveillance.6 In the setting of public reporting and nonpayment, the method of identifying CDAD cases must be accurate to ensure fairness, while being inexpensive. To identify the potential value of these less labor‐intensive methods of reporting the incidence of CDAD, we evaluated the use of ICD‐9 codes and C. difficile toxin to accurately report the incidence of CDAD at a community hospital, as well as the labor hours required for each reporting method.
METHODS
Patients >18 years of age and potentially having CDAD were identified via database queries for ICD‐9 codes and positive C. difficile toxin assays at our institution from November 1, 2006, through August 31, 2007. Our institution is a 379‐bed university‐affiliated community teaching hospital in a socio‐economically diverse area of Baltimorewith approximately 110,000 emergency department visits, 30,000 discharges, and 110,000 inpatient days each yearthat uses a handwritten paper chart for all provider orders and patient documentation. The C. difficile toxin assay method used during the study period was an enzyme immunoassay that detects both toxins A and B (Meridan Bioscience, Cincinnati, OH). ICD‐9 codes were queried in the CareScience database(Premier, Inc; Charlotte, NC), while C. difficile toxin was queried via the hospital laboratory database. All identified patients underwent a medical record review to confirm the diagnosis of CDAD and the patient's location at the time of disease acquisition. To eliminate duplicate reporting of a single episode of disease, all duplicate patient visits with a diagnosis of CDAD within 30 days were removed.
Our diagnostic criteria (Table 1) used the recommended criteria of a combination of symptoms and positive toxin assay, visualization of pseudomembranes on colonoscopy or pathology‐proven CDAD.6 Based on the presence of CDAD in 25% of patients with a white blood cell count >30,000 and the recommendation that CDAD be considered in all patients with a white blood cell count >15,000, we included the criterion of clinical findings with severe leukocytosis to identify patients with toxin‐negative CDAD.14
| Chart Review Criteria for CDAD | Chart Review Criteria for CDAD Obtained While Patient Was Not at Our Hospital | Chart Review Criteria for CDAD Obtained While Patient Was at Our Hospital |
|---|---|---|
| ||
| Pseudomembranous colitis seen during endoscopy OR biopsy or resection with surgical pathology consistent with CDAD | One of the above symptoms or objective criteria positive within the first 3 days of hospitalization AND patient was not cared for at our insitution within the last 7 days | Patient was cared for at our institution within 7 days of presentation OR the patient's symptoms were not present upon arrival at the hospital and the symptoms began after the third day of hospitalization |
| If neither of the above are present, then Clostridium difficile toxin or WBC count >25,000 plus one of the following: | ||
| Diarrhea | ||
| Fever without other cause | ||
| Abdominal pain without other cause | ||
| Colonic ileus without other cause | ||
Patients were identified by querying the CareScience database for patients having an ICD‐9 code of 008.45. Duplicate cases for the same patient within 30 days were removed. These cases were compared with the cases of CDAD determined by the medical record review for analysis. Patients were identified via positive C. difficile toxin by querying the microbiology laboratory database. A query of all patients with a positive C. difficile toxin was performed. Duplicate samples for the same patient within 30 days were removed. Patients are considered to be outside the hospital when CDAD was acquired if their toxin was positive within the first 3 days of arrival to the hospital, and they were not hospitalized at our institution within 1 week before arrival. All other cases were considered to be acquired while in our hospital. The number of patients with a toxin‐positive stool sample was compared with the medical record review.
Recognizing that it is unrealistic to review the medical records of the 23,495 discharges during the 10‐month study period, a random sample review of 500 charts not included in our CDAD‐identified patient list was performed to identify the rate at which CDAD patients failed to be identified by our methods. From a list of discharges during the study period, as long as they were not in our study population, every thirtieth patient was selected for review up to a total of 500 patient charts. Then, the identified case rate was used to predict the prevalence of disease at our institution during the study period.
Sample selection was based on the absence of previous studies evaluating the performance of C. difficile toxin assay, and the desire to have the same assay used during the entire study. The lack of data from which to perform sample size determination would result in an inaccurate estimate. Therefore, we chose a period that offered the maximum sample size, during which a single assay was used at our institution. Compared with medical record review, the accuracy and positive predictive values of the ICD‐9 code and positive stool toxin methods to identify the cases of CDAD were compared. A true positive was when the ICD‐9 or laboratory query method identified a patient who had CDAD based on chart review; a false positive was when the ICD‐9 or laboratory query method identified a patient who did not have CDAD based on chart review. Rates of over‐ or underdiagnosis, case rates, and acquisition location were determined. Statistical analysis of the case rates and acquisition location were performed via chi‐square test. The time to perform these queries was collected with accuracy to the minute.
RESULTS
Of the 23,495 discharges during the study period, the combination of ICD‐9 and C. difficile toxin assay identified a total cohort of 496 patients, 319 of whom were identified by both the ICD‐9 method and the toxin assay, 50 of whom were identified only by the toxin assay, and 127 of whom were identified only by the ICD‐9 method. Chart review confirmed the presence of CDAD in 384 of these 496 cases, for a case rate of 16.3 per 1000 discharged patients (Table 2). The diagnostic criteria for each of these confirmed cases are listed in Table 3. Of the 384 confirmed CDAD cases, 319 were identified by both the ICD‐9 and toxin assay, 50 were identified only by the toxin assay, and 15 were identified only by the ICD‐9 query. Of the 50 cases identified by the toxin assay that were not identified via the ICD‐9 method, all 50 (100%) were confirmed to have CDAD by chart review. In contrast, of the 127 cases identified via the ICD‐9 method that were not found via the toxin assay, only 15 (11.8%) were confirmed to have CDAD by chart review (Figure 1).

| ICD‐9 | Toxin Assay | Chart Review | |
|---|---|---|---|
| |||
| No. of patients identified | 446 | 369 | 384 |
| Case rate per 1000 discharges* | 19.0 | 15.7 | 16.3 |
| 95% confidence interval | 17.320.8 | 14.117.4 | NA |
| Case rate compared with chart review, P | P = 0.001 | P = 0.440 | NA |
| CDAD rate reported compared with chart review | 116.1% | 96.1% | NA |
| Accuracy | 83.9% | 96.1% | NA |
| PPV | 74.9% | 100% | NA |
| Portion of cases acquired at our hospital, % | NA | 48.2% | 40.6% |
| Portion of cases acquired at our hospital, P | NA | P = 0.003 | NA |
| Minutes consumed for data collection | 312 | 842 | 21,899 |
| Estimated annual cost per hospital | $234.00 | $631.50 | $16,424.25 |
| Criterion | No. of Cases | Case Rate* |
|---|---|---|
| ||
| Endoscopy | 2 | 0.5% |
| Surgical pathology | 9 | 2.3% |
| Positive toxin and diarrhea | 369 | 96.1% |
| WBC >25.0 and diarrhea | 51 | 13.3% |
| WBC >25.0 and fever without other source | 9 | 2.3% |
| WBC >25.0 and unexplained abdominal pain | 17 | 4.4% |
| WBC >25.0 and colonic ileus | 2 | 0.5% |
Of these 384 cases, 369 were identified via the toxin assay for a case rate of 15.7 per 1000 patient discharges, which was not found to be different from the rate of 16.3 determined by chart review (P = 0.440; 95% confidence interval [CI], 14.117.4). Compared with chart review, the toxin assay reported 96.1% (369/384) of cases. Chart review demonstrated that every patient who had a positive toxin assay met the diagnostic criteria for CDAD for a positive predictive value (PPV) of 100%.
The ICD‐9 method identified 446 patients thought to have CDAD, 334 of whom were confirmed by chart review for a PPV of 74.9% (334/446). Compared with chart review, the ICD‐9 method reported 116.1% (446/384) of cases for a case rate of 19.0 per 1000 discharges and was significantly different than the rate of 16.3 reported by chart review (P = 0.001; 95% CI, 17.320.8).
Chart review identified 156 of 384 (40.6%) patients who acquired CDAD while at our hospital and 228 of 384 (59.4%) who acquired it elsewhere. In comparison, the toxin assay criteria identified 369 cases of CDAD, of which 48.2% (178/369) were acquired while at our hospital and 51.8% (191/369) were acquired elsewhere (P = 0.003).
The time for data extraction via these 3 methods differed greatly. The ICD‐9 method only consumed 312 minutes and the toxin assay method 842 minutes, whereas the chart review method consumed 21,899 minutes. These times reported include the database query and data analysis for the ICD‐9 and toxin assay methods, while it includes the database query and list generation along with the manual chart review and data analysis for the chart review method. The review of the random sample of patients believed not to have CDAD was not included in any of the reported times. Chart review on a random sample of 500 patients not previously identified for review found no additional cases of CDAD.
DISCUSSION
Our study demonstrates that use of positive C. difficile toxin assay data from the microbiology laboratory alone is an efficient method of identifying patients with CDAD. This method only consumed 842 minutes versus 21,899 minutes consumed by the chart review method. The C. difficile toxin method reduces the workforce required to collect and analyze this data, but more importantly, it was found to be reliable by reporting an institutional case rate that is similar to that of chart review.
In contrast, the ICD‐9 method was efficient but less reliable. It only consumed 312 minutes, but it overreported the institutional rate of CDAD by 16.1%, had a PPV of only 74.9%, and of those patients who were identified by ICD‐9 but not toxin assay, only 11.8% actually had CDAD. This finding is in conflict with the previously noted underreporting of this method.13, 15 We believe this difference to be associated with institutional differences, because previous reports originated from a veterans hospital and an academic medical center, and previous authors have failed to use predefined diagnostic criteria and a complete chart review to confirm cases of CDAD. Similar to our study, an academic medical center identified that listing of CDAD in a patient's medical history in their chart was associated with a false‐positive ICD‐9 code for CDAD.15 This observation appears to bring clarity to one of the causes of the variance of ICD‐9 code accuracy between institutions. Institutions seem to vary on the method of attaching diagnoses to the patient's final hospital record. Some institutions include only what is listed as an active problem, whereas others list diagnoses listed in the chart as previous problems and those listed as a potential diagnosis without confirmation. Another potential cause of the ICD‐9 inaccuracy is the potential of clinicians to diagnose and treat a patient for CDAD in the absence of the diagnostic criteria used for chart review. Physician practices such as these are known to vary between institutions leading to a variance in the ICD‐9 code accuracy.
In total, 15 cases of CDAD were identified in the absence of a positive toxin assay, and of these, 12 cases were identified using leukocytosis‐based criteria (Table 4). This resulted in 3.1% of our cases being toxin‐negative, based on leukocytosis criteria, and is lower than the previously identified 35% of cases.14 Because it was the only assay available at the time, this previous research used a toxin Aonly assay, which is more likely to have false‐negative results than the toxin A/B assay used at our institution during the study period. The investigators also required all toxin‐negative patients to have been recently treated with antibiotics. Based on the increasing rates of community‐acquired CDAD, including those that are antibiotic‐nave patients, we felt a history of antibiotic exposure was no longer a prerequisite for CDAD and thus excluded it from our diagnostic criteria.16, 17 Based on these differences, we feel our results are likely an accurate reflection of the number of cases identified by ICD‐9 query in the absence of toxin positivity. However, concerns should be further alleviated through the realization that nonuse of this strategy improves the accuracy of the toxin assay method, while reducing the accuracy of the ICD‐9 method and thereby strengthening the validity of our conclusions. Mathematically, this would result in 369 of 369 patients identified by toxin assay and 369 patients identified by the ICD‐9 method. This would reduce the case rate of the chart review method to the same 15.7 of the toxin assay, while the ICD‐9 method would remain at 19.0.
| Criterion | No. of Cases* |
|---|---|
| |
| Endoscopy | 0 |
| Surgical pathology | 3 |
| WBC >25.0 and diarrhea | 10 |
| WBC >25.0 and fever without other source | 1 |
| WBC >25.0 and unexplained abdominal pain | 3 |
| WBC >25.0 and colonic ileus | 1 |
We considered whether the toxin assay method may overestimate the number of cases due to a C. difficileasymptomatic carrier rate as high as 50% of hospitalized patients.6 However, we found no difference in the case rate when compared with that of chart review, and there were no false‐positive cases. We believe this is attributable to the 30‐day window that was used to identify a single episode of CDAD and the absence of toxin assays being performed on asymptomatic individuals. To avoid overrepresentation of the actual number of CDAD cases, we chose to label all repeat positive toxins and repeat hospitalizations within 30 days as a single episode of CDAD. This was based on the identification that 56% of patients remained toxin‐positive 26 weeks after adequate treatment for CDAD.18 The enzyme‐linked immunosorbent assay (ELISA) method used by our laboratory is the method used to report 94% of CDAD cases in a national point prevalence study that collected data from United States acute care hospitals with representation from 47 states1, 6, 19 (Table 5). This use of ELISA in the majority of United States hospitals suggests that our data can be extrapolated for use throughout the United States. While laboratories are increasing their use of 2‐step algorithms involving glutamate dehydrogenase antigen assay followed by cytotoxin neutralization, and more recently beginning the use of polymerase chain reaction assays, both of these methods have been found to increase the accuracy of detecting C. difficile compared with ELISA.20 Therefore, as laboratories evolve to use more accurate assays to detect CDAD, the methods described herein will be expected to increase in reliability.
| Method | Sensitivity | Specificity | Cost | Ease of Performance | Typical Results Reporting | Notes |
|---|---|---|---|---|---|---|
| ||||||
| Culture | Gold standard | NA | $$$$ | Difficult | Days | Slow turnaround time; is the standard upon which other test methods are based, but not all organisms are toxin‐producing |
| Cell cytotoxicity assay | 67%100% | Gold standard | $$$ | Intermediate | Next day | Is the standard upon which other test methods are based to identify toxin‐producing stains of Clostridium difficile |
| EIA for toxin A/B | 63%94% | 75%100% | $ | Easy | Same day | Used by >90% of laboratories in the United States |
| EIA for detection of Clostridium difficile common antigen (GDH) | 85%95% | 89%99% | $ | Easy | Same day | Provides no information regarding the toxigenicity of the isolate, typically used in combination with cell cytotoxicity assay to identify toxin‐producing strains |
| Polymerase chain reaction | 96%100% | 88%91% | $$ | Intermediate | Same day | More data are needed before recommendation for routine testing |
The toxin assay methodology used to determine the rate of CDAD cases acquired while the patient was at our hospital overreported these cases. Based on this result, identification of individual cases of CDAD that are obtained at a specific hospital would continue to require manual chart review. This expensive method may be avoided by instead choosing to use institutional case rates for reporting, monitoring, and incentivizing hospitals. However, a discussion of the methods of this approach and its confluence with our societal goal to move toward Accountable Care Organizations is beyond the scope of this discussion section.
Although it appears that we identified all cases of CDAD occurring at our institution, a limitation of this study is its inability to review all charts during the 10‐month study period. We used a combination of ICD‐9 and positive C. difficile toxin assay data to identify all possible cases of C. difficile. The current approach to case identification for reported hospital conditions is limited to an ICD‐9 database query. This query is followed by chart review to collect data for hospital performance that is published in locations such as
Increased automation is expected in the future of reporting. The Centers for Disease Control and Prevention found increased rates of disease reporting and increased accuracy when reporting is electronically automated via their software system, Electronic Support for Public Health, which is designed to communicate with and perform automated data queries on providers' electronic medical records.21 While use of this model is creeping into the health system for reporting to public health authorities,22 universal hospital electronic medical record implementation and full connectivity with such reporting systems is many years from fruition. In addition to its practical use for reporting CDAD in our current health system, our work easily transitions into automated reporting within an electronically integrated health system once achieved.
In conclusion, ICD‐9 data were found to be unreliable, and consideration must be given to cessation of their use for CDAD case rate research and reporting. Use of a positive C. difficile toxin assay accurately reports the institutional incidence of disease, can be used by individual institutions to self‐monitor case rates or by the government to determine regionally acceptable intuitional rates of CDAD on which incentives and penalties can be based, and will increase in efficiency as reporting continues to be automated. This process can be instituted at a fraction of the cost of the standard chart review that is currently used for most reporting.
With an increased incidence of 13.1 per 1000 inpatients1, 2 and an attributable mortality of 6.1%,3 in 2006 the Canadian government added Clostridium difficileassociated disease (CDAD) to its list of reportable diseases.4 The Centers for Disease Control and Prevention and the Infectious Disease Society of America subsequently created definitions of the disease and recommended surveillance of rates of health care facilityassociated CDAD, which has been found to double a patient's length of stay and cost of hospitalization.57 While not included in the current list of hospital‐acquired conditions for which payment is declined,8 the Centers for Medicare & Medicaid Services (CMS) has noted CDAD as under consideration for addition to the list,9 which would require hospital reporting of CDAD rates. This reporting is typically a labor‐intensive, medical record review process that increases the cost of delivering health care. Health care institutions have reported spending as much as $21 million per year or $400 per discharged patient on quality improvement.10 These costs are passed on to payers such as those governed by the CMS that are projected to pay for more than half of all national health spending in 2018.11 Therefore, it is prudent to examine alternatives for determining rates of disease for public reporting and quality improvement initiatives such as infection control and antibiotic stewardship programs.
There are 3 methods that can be used for this reporting. First, medical record review has been used for determining the case rate of CDAD by published reports.1, 2 This procedure allows the CMS to define the desired data to be collected, but it is labor‐intensive. Second, the use of International Statistical Classification of Diseases and Related Health Problems, 9th Edition (ICD‐9) codes from hospital databases offers the speed of database query. However, due to diagnostic and coding errors, it may report inaccurate rates of disease. Previous reports include a university‐based hospital that found a sensitivity of 78% and specificity of 99.7%,12 whereas a Veterans Administration medical center found nearly two‐thirds of patients with CDAD did not have the ICD‐9 code for C. difficile infection noted in their database.13 Therefore, the accuracy of ICD‐9 code at a community hospital that better represents the majority of United States hospitals is needed. The third method of identifying CDAD is the presence of toxin identified in the microbiology laboratory. Although this method offers ease of obtainment, it suffers from potential inaccuracy arising from duplicate patient samples, positive samples in patients who are symptom‐free carriers, and patients with CDAD despite having a negative toxin.
The same organizations that recommend surveillance for CDAD have identified that there are inadequate data on which to base a decision regarding how to proceed with routine community and hospital surveillance.6 In the setting of public reporting and nonpayment, the method of identifying CDAD cases must be accurate to ensure fairness, while being inexpensive. To identify the potential value of these less labor‐intensive methods of reporting the incidence of CDAD, we evaluated the use of ICD‐9 codes and C. difficile toxin to accurately report the incidence of CDAD at a community hospital, as well as the labor hours required for each reporting method.
METHODS
Patients >18 years of age and potentially having CDAD were identified via database queries for ICD‐9 codes and positive C. difficile toxin assays at our institution from November 1, 2006, through August 31, 2007. Our institution is a 379‐bed university‐affiliated community teaching hospital in a socio‐economically diverse area of Baltimorewith approximately 110,000 emergency department visits, 30,000 discharges, and 110,000 inpatient days each yearthat uses a handwritten paper chart for all provider orders and patient documentation. The C. difficile toxin assay method used during the study period was an enzyme immunoassay that detects both toxins A and B (Meridan Bioscience, Cincinnati, OH). ICD‐9 codes were queried in the CareScience database(Premier, Inc; Charlotte, NC), while C. difficile toxin was queried via the hospital laboratory database. All identified patients underwent a medical record review to confirm the diagnosis of CDAD and the patient's location at the time of disease acquisition. To eliminate duplicate reporting of a single episode of disease, all duplicate patient visits with a diagnosis of CDAD within 30 days were removed.
Our diagnostic criteria (Table 1) used the recommended criteria of a combination of symptoms and positive toxin assay, visualization of pseudomembranes on colonoscopy or pathology‐proven CDAD.6 Based on the presence of CDAD in 25% of patients with a white blood cell count >30,000 and the recommendation that CDAD be considered in all patients with a white blood cell count >15,000, we included the criterion of clinical findings with severe leukocytosis to identify patients with toxin‐negative CDAD.14
| Chart Review Criteria for CDAD | Chart Review Criteria for CDAD Obtained While Patient Was Not at Our Hospital | Chart Review Criteria for CDAD Obtained While Patient Was at Our Hospital |
|---|---|---|
| ||
| Pseudomembranous colitis seen during endoscopy OR biopsy or resection with surgical pathology consistent with CDAD | One of the above symptoms or objective criteria positive within the first 3 days of hospitalization AND patient was not cared for at our insitution within the last 7 days | Patient was cared for at our institution within 7 days of presentation OR the patient's symptoms were not present upon arrival at the hospital and the symptoms began after the third day of hospitalization |
| If neither of the above are present, then Clostridium difficile toxin or WBC count >25,000 plus one of the following: | ||
| Diarrhea | ||
| Fever without other cause | ||
| Abdominal pain without other cause | ||
| Colonic ileus without other cause | ||
Patients were identified by querying the CareScience database for patients having an ICD‐9 code of 008.45. Duplicate cases for the same patient within 30 days were removed. These cases were compared with the cases of CDAD determined by the medical record review for analysis. Patients were identified via positive C. difficile toxin by querying the microbiology laboratory database. A query of all patients with a positive C. difficile toxin was performed. Duplicate samples for the same patient within 30 days were removed. Patients are considered to be outside the hospital when CDAD was acquired if their toxin was positive within the first 3 days of arrival to the hospital, and they were not hospitalized at our institution within 1 week before arrival. All other cases were considered to be acquired while in our hospital. The number of patients with a toxin‐positive stool sample was compared with the medical record review.
Recognizing that it is unrealistic to review the medical records of the 23,495 discharges during the 10‐month study period, a random sample review of 500 charts not included in our CDAD‐identified patient list was performed to identify the rate at which CDAD patients failed to be identified by our methods. From a list of discharges during the study period, as long as they were not in our study population, every thirtieth patient was selected for review up to a total of 500 patient charts. Then, the identified case rate was used to predict the prevalence of disease at our institution during the study period.
Sample selection was based on the absence of previous studies evaluating the performance of C. difficile toxin assay, and the desire to have the same assay used during the entire study. The lack of data from which to perform sample size determination would result in an inaccurate estimate. Therefore, we chose a period that offered the maximum sample size, during which a single assay was used at our institution. Compared with medical record review, the accuracy and positive predictive values of the ICD‐9 code and positive stool toxin methods to identify the cases of CDAD were compared. A true positive was when the ICD‐9 or laboratory query method identified a patient who had CDAD based on chart review; a false positive was when the ICD‐9 or laboratory query method identified a patient who did not have CDAD based on chart review. Rates of over‐ or underdiagnosis, case rates, and acquisition location were determined. Statistical analysis of the case rates and acquisition location were performed via chi‐square test. The time to perform these queries was collected with accuracy to the minute.
RESULTS
Of the 23,495 discharges during the study period, the combination of ICD‐9 and C. difficile toxin assay identified a total cohort of 496 patients, 319 of whom were identified by both the ICD‐9 method and the toxin assay, 50 of whom were identified only by the toxin assay, and 127 of whom were identified only by the ICD‐9 method. Chart review confirmed the presence of CDAD in 384 of these 496 cases, for a case rate of 16.3 per 1000 discharged patients (Table 2). The diagnostic criteria for each of these confirmed cases are listed in Table 3. Of the 384 confirmed CDAD cases, 319 were identified by both the ICD‐9 and toxin assay, 50 were identified only by the toxin assay, and 15 were identified only by the ICD‐9 query. Of the 50 cases identified by the toxin assay that were not identified via the ICD‐9 method, all 50 (100%) were confirmed to have CDAD by chart review. In contrast, of the 127 cases identified via the ICD‐9 method that were not found via the toxin assay, only 15 (11.8%) were confirmed to have CDAD by chart review (Figure 1).

| ICD‐9 | Toxin Assay | Chart Review | |
|---|---|---|---|
| |||
| No. of patients identified | 446 | 369 | 384 |
| Case rate per 1000 discharges* | 19.0 | 15.7 | 16.3 |
| 95% confidence interval | 17.320.8 | 14.117.4 | NA |
| Case rate compared with chart review, P | P = 0.001 | P = 0.440 | NA |
| CDAD rate reported compared with chart review | 116.1% | 96.1% | NA |
| Accuracy | 83.9% | 96.1% | NA |
| PPV | 74.9% | 100% | NA |
| Portion of cases acquired at our hospital, % | NA | 48.2% | 40.6% |
| Portion of cases acquired at our hospital, P | NA | P = 0.003 | NA |
| Minutes consumed for data collection | 312 | 842 | 21,899 |
| Estimated annual cost per hospital | $234.00 | $631.50 | $16,424.25 |
| Criterion | No. of Cases | Case Rate* |
|---|---|---|
| ||
| Endoscopy | 2 | 0.5% |
| Surgical pathology | 9 | 2.3% |
| Positive toxin and diarrhea | 369 | 96.1% |
| WBC >25.0 and diarrhea | 51 | 13.3% |
| WBC >25.0 and fever without other source | 9 | 2.3% |
| WBC >25.0 and unexplained abdominal pain | 17 | 4.4% |
| WBC >25.0 and colonic ileus | 2 | 0.5% |
Of these 384 cases, 369 were identified via the toxin assay for a case rate of 15.7 per 1000 patient discharges, which was not found to be different from the rate of 16.3 determined by chart review (P = 0.440; 95% confidence interval [CI], 14.117.4). Compared with chart review, the toxin assay reported 96.1% (369/384) of cases. Chart review demonstrated that every patient who had a positive toxin assay met the diagnostic criteria for CDAD for a positive predictive value (PPV) of 100%.
The ICD‐9 method identified 446 patients thought to have CDAD, 334 of whom were confirmed by chart review for a PPV of 74.9% (334/446). Compared with chart review, the ICD‐9 method reported 116.1% (446/384) of cases for a case rate of 19.0 per 1000 discharges and was significantly different than the rate of 16.3 reported by chart review (P = 0.001; 95% CI, 17.320.8).
Chart review identified 156 of 384 (40.6%) patients who acquired CDAD while at our hospital and 228 of 384 (59.4%) who acquired it elsewhere. In comparison, the toxin assay criteria identified 369 cases of CDAD, of which 48.2% (178/369) were acquired while at our hospital and 51.8% (191/369) were acquired elsewhere (P = 0.003).
The time for data extraction via these 3 methods differed greatly. The ICD‐9 method only consumed 312 minutes and the toxin assay method 842 minutes, whereas the chart review method consumed 21,899 minutes. These times reported include the database query and data analysis for the ICD‐9 and toxin assay methods, while it includes the database query and list generation along with the manual chart review and data analysis for the chart review method. The review of the random sample of patients believed not to have CDAD was not included in any of the reported times. Chart review on a random sample of 500 patients not previously identified for review found no additional cases of CDAD.
DISCUSSION
Our study demonstrates that use of positive C. difficile toxin assay data from the microbiology laboratory alone is an efficient method of identifying patients with CDAD. This method only consumed 842 minutes versus 21,899 minutes consumed by the chart review method. The C. difficile toxin method reduces the workforce required to collect and analyze this data, but more importantly, it was found to be reliable by reporting an institutional case rate that is similar to that of chart review.
In contrast, the ICD‐9 method was efficient but less reliable. It only consumed 312 minutes, but it overreported the institutional rate of CDAD by 16.1%, had a PPV of only 74.9%, and of those patients who were identified by ICD‐9 but not toxin assay, only 11.8% actually had CDAD. This finding is in conflict with the previously noted underreporting of this method.13, 15 We believe this difference to be associated with institutional differences, because previous reports originated from a veterans hospital and an academic medical center, and previous authors have failed to use predefined diagnostic criteria and a complete chart review to confirm cases of CDAD. Similar to our study, an academic medical center identified that listing of CDAD in a patient's medical history in their chart was associated with a false‐positive ICD‐9 code for CDAD.15 This observation appears to bring clarity to one of the causes of the variance of ICD‐9 code accuracy between institutions. Institutions seem to vary on the method of attaching diagnoses to the patient's final hospital record. Some institutions include only what is listed as an active problem, whereas others list diagnoses listed in the chart as previous problems and those listed as a potential diagnosis without confirmation. Another potential cause of the ICD‐9 inaccuracy is the potential of clinicians to diagnose and treat a patient for CDAD in the absence of the diagnostic criteria used for chart review. Physician practices such as these are known to vary between institutions leading to a variance in the ICD‐9 code accuracy.
In total, 15 cases of CDAD were identified in the absence of a positive toxin assay, and of these, 12 cases were identified using leukocytosis‐based criteria (Table 4). This resulted in 3.1% of our cases being toxin‐negative, based on leukocytosis criteria, and is lower than the previously identified 35% of cases.14 Because it was the only assay available at the time, this previous research used a toxin Aonly assay, which is more likely to have false‐negative results than the toxin A/B assay used at our institution during the study period. The investigators also required all toxin‐negative patients to have been recently treated with antibiotics. Based on the increasing rates of community‐acquired CDAD, including those that are antibiotic‐nave patients, we felt a history of antibiotic exposure was no longer a prerequisite for CDAD and thus excluded it from our diagnostic criteria.16, 17 Based on these differences, we feel our results are likely an accurate reflection of the number of cases identified by ICD‐9 query in the absence of toxin positivity. However, concerns should be further alleviated through the realization that nonuse of this strategy improves the accuracy of the toxin assay method, while reducing the accuracy of the ICD‐9 method and thereby strengthening the validity of our conclusions. Mathematically, this would result in 369 of 369 patients identified by toxin assay and 369 patients identified by the ICD‐9 method. This would reduce the case rate of the chart review method to the same 15.7 of the toxin assay, while the ICD‐9 method would remain at 19.0.
| Criterion | No. of Cases* |
|---|---|
| |
| Endoscopy | 0 |
| Surgical pathology | 3 |
| WBC >25.0 and diarrhea | 10 |
| WBC >25.0 and fever without other source | 1 |
| WBC >25.0 and unexplained abdominal pain | 3 |
| WBC >25.0 and colonic ileus | 1 |
We considered whether the toxin assay method may overestimate the number of cases due to a C. difficileasymptomatic carrier rate as high as 50% of hospitalized patients.6 However, we found no difference in the case rate when compared with that of chart review, and there were no false‐positive cases. We believe this is attributable to the 30‐day window that was used to identify a single episode of CDAD and the absence of toxin assays being performed on asymptomatic individuals. To avoid overrepresentation of the actual number of CDAD cases, we chose to label all repeat positive toxins and repeat hospitalizations within 30 days as a single episode of CDAD. This was based on the identification that 56% of patients remained toxin‐positive 26 weeks after adequate treatment for CDAD.18 The enzyme‐linked immunosorbent assay (ELISA) method used by our laboratory is the method used to report 94% of CDAD cases in a national point prevalence study that collected data from United States acute care hospitals with representation from 47 states1, 6, 19 (Table 5). This use of ELISA in the majority of United States hospitals suggests that our data can be extrapolated for use throughout the United States. While laboratories are increasing their use of 2‐step algorithms involving glutamate dehydrogenase antigen assay followed by cytotoxin neutralization, and more recently beginning the use of polymerase chain reaction assays, both of these methods have been found to increase the accuracy of detecting C. difficile compared with ELISA.20 Therefore, as laboratories evolve to use more accurate assays to detect CDAD, the methods described herein will be expected to increase in reliability.
| Method | Sensitivity | Specificity | Cost | Ease of Performance | Typical Results Reporting | Notes |
|---|---|---|---|---|---|---|
| ||||||
| Culture | Gold standard | NA | $$$$ | Difficult | Days | Slow turnaround time; is the standard upon which other test methods are based, but not all organisms are toxin‐producing |
| Cell cytotoxicity assay | 67%100% | Gold standard | $$$ | Intermediate | Next day | Is the standard upon which other test methods are based to identify toxin‐producing stains of Clostridium difficile |
| EIA for toxin A/B | 63%94% | 75%100% | $ | Easy | Same day | Used by >90% of laboratories in the United States |
| EIA for detection of Clostridium difficile common antigen (GDH) | 85%95% | 89%99% | $ | Easy | Same day | Provides no information regarding the toxigenicity of the isolate, typically used in combination with cell cytotoxicity assay to identify toxin‐producing strains |
| Polymerase chain reaction | 96%100% | 88%91% | $$ | Intermediate | Same day | More data are needed before recommendation for routine testing |
The toxin assay methodology used to determine the rate of CDAD cases acquired while the patient was at our hospital overreported these cases. Based on this result, identification of individual cases of CDAD that are obtained at a specific hospital would continue to require manual chart review. This expensive method may be avoided by instead choosing to use institutional case rates for reporting, monitoring, and incentivizing hospitals. However, a discussion of the methods of this approach and its confluence with our societal goal to move toward Accountable Care Organizations is beyond the scope of this discussion section.
Although it appears that we identified all cases of CDAD occurring at our institution, a limitation of this study is its inability to review all charts during the 10‐month study period. We used a combination of ICD‐9 and positive C. difficile toxin assay data to identify all possible cases of C. difficile. The current approach to case identification for reported hospital conditions is limited to an ICD‐9 database query. This query is followed by chart review to collect data for hospital performance that is published in locations such as
Increased automation is expected in the future of reporting. The Centers for Disease Control and Prevention found increased rates of disease reporting and increased accuracy when reporting is electronically automated via their software system, Electronic Support for Public Health, which is designed to communicate with and perform automated data queries on providers' electronic medical records.21 While use of this model is creeping into the health system for reporting to public health authorities,22 universal hospital electronic medical record implementation and full connectivity with such reporting systems is many years from fruition. In addition to its practical use for reporting CDAD in our current health system, our work easily transitions into automated reporting within an electronically integrated health system once achieved.
In conclusion, ICD‐9 data were found to be unreliable, and consideration must be given to cessation of their use for CDAD case rate research and reporting. Use of a positive C. difficile toxin assay accurately reports the institutional incidence of disease, can be used by individual institutions to self‐monitor case rates or by the government to determine regionally acceptable intuitional rates of CDAD on which incentives and penalties can be based, and will increase in efficiency as reporting continues to be automated. This process can be instituted at a fraction of the cost of the standard chart review that is currently used for most reporting.
- ,,,.National point prevalence of Clostridium difficile in US health care facility inpatients, 2008.Am J Infect Control.2009;37:263–270.
- ,,.Secular trends in hospital‐acquired Clostridium difficile disease in the United States, 1987–2001.J Infect Dis.2004;189:1585–1589.
- ,,, et al.Analysis of 30‐day mortality for Clostridium difficile‐associated disease in the ICU setting.Chest.2007;132:418–424.
- .Final report and recommendations from the National Notifiable Diseases Working Group.Can Commun Dis Rep.2006;32:211–225.
- ,,, et al.Recommendations for surveillance of Clostridium difficile‐associated disease.Infect Control Hosp Epidemiol.2007;28:140–145.
- ,,, et al.Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA).Infect Control Hosp Epidemiol.2010;31:431–455.
- ,.Review of current literature on the economic burden of Clostridium difficile infection.Infect Control Hosp Epidemiol.2009;30:57–66.
- Centers for Medicare 35:544–550.
- ,,, et al.Health spending projections through 2018: recession effects add uncertainty to the outlook.Health Aff (Millwood).2009;28:w346–w357.
- ,,,.ICD‐9 codes and surveillance for Clostridium difficile‐associated disease.Emerg Infect Dis.2006;12:1576–1579.
- ,,,.Fluoroquinolone use and risk factors for Clostridium difficile‐associated disease within a Veterans Administration health care system.Clin Infect Dis.2007;45:1141–1151.
- ,,.Conditions associated with leukocytosis in a tertiary care hospital, with particular attention to the role of infection caused by clostridium difficile.Clin Infect Dis.2002;34:1585–1592.
- ,,,.Accuracy of ICD‐9 coding for Clostridium difficile infections: a retrospective cohort.Epidemiol Infect.2007;135:1010–1013.
- ,,,.Implications of the changing face of Clostridium difficile disease for health care practitioners.Am J Infect Control.2007;35:237–253.
- .Clostridium difficile is no longer just a nosocomial infection or an infection of adults.Int J Antimicrob Agents.2009;33(suppl 1):S42–S45.
- ,,,,.Treatment of antibiotic‐associated Clostridium difficile colitis with oral vancomycin: comparison of two dosage regimens.Am J Med.1989;86:15–19.
- ,,, et al.Comparison of real‐time PCR techniques to cytotoxigenic culture methods for diagnosing Clostridium difficile infection.J Clin Microbiol.2011;49:227–231.
- ,,,.Comparison of BD GeneOhm Cdiff real‐time PCR assay with a two‐step algorithm and a toxin A/B enzyme‐linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection.J Clin Microbiol.2010;48:109–114.
- Centers for Disease Control and Prevention.Automated detection and reporting of notifiable diseases using electronic medical records versus passive surveillance—Massachusetts, June 2006‐July 2007.MMWR Morb Mortal Wkly Rep.2008;57:373–376.
- ,,, et al.Development of an electronic public health case report using HL7 v2.5 to meet public health needs.J Am Med Inform Assoc.2010;17:34–41.
- ,,,.National point prevalence of Clostridium difficile in US health care facility inpatients, 2008.Am J Infect Control.2009;37:263–270.
- ,,.Secular trends in hospital‐acquired Clostridium difficile disease in the United States, 1987–2001.J Infect Dis.2004;189:1585–1589.
- ,,, et al.Analysis of 30‐day mortality for Clostridium difficile‐associated disease in the ICU setting.Chest.2007;132:418–424.
- .Final report and recommendations from the National Notifiable Diseases Working Group.Can Commun Dis Rep.2006;32:211–225.
- ,,, et al.Recommendations for surveillance of Clostridium difficile‐associated disease.Infect Control Hosp Epidemiol.2007;28:140–145.
- ,,, et al.Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA).Infect Control Hosp Epidemiol.2010;31:431–455.
- ,.Review of current literature on the economic burden of Clostridium difficile infection.Infect Control Hosp Epidemiol.2009;30:57–66.
- Centers for Medicare 35:544–550.
- ,,, et al.Health spending projections through 2018: recession effects add uncertainty to the outlook.Health Aff (Millwood).2009;28:w346–w357.
- ,,,.ICD‐9 codes and surveillance for Clostridium difficile‐associated disease.Emerg Infect Dis.2006;12:1576–1579.
- ,,,.Fluoroquinolone use and risk factors for Clostridium difficile‐associated disease within a Veterans Administration health care system.Clin Infect Dis.2007;45:1141–1151.
- ,,.Conditions associated with leukocytosis in a tertiary care hospital, with particular attention to the role of infection caused by clostridium difficile.Clin Infect Dis.2002;34:1585–1592.
- ,,,.Accuracy of ICD‐9 coding for Clostridium difficile infections: a retrospective cohort.Epidemiol Infect.2007;135:1010–1013.
- ,,,.Implications of the changing face of Clostridium difficile disease for health care practitioners.Am J Infect Control.2007;35:237–253.
- .Clostridium difficile is no longer just a nosocomial infection or an infection of adults.Int J Antimicrob Agents.2009;33(suppl 1):S42–S45.
- ,,,,.Treatment of antibiotic‐associated Clostridium difficile colitis with oral vancomycin: comparison of two dosage regimens.Am J Med.1989;86:15–19.
- ,,, et al.Comparison of real‐time PCR techniques to cytotoxigenic culture methods for diagnosing Clostridium difficile infection.J Clin Microbiol.2011;49:227–231.
- ,,,.Comparison of BD GeneOhm Cdiff real‐time PCR assay with a two‐step algorithm and a toxin A/B enzyme‐linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection.J Clin Microbiol.2010;48:109–114.
- Centers for Disease Control and Prevention.Automated detection and reporting of notifiable diseases using electronic medical records versus passive surveillance—Massachusetts, June 2006‐July 2007.MMWR Morb Mortal Wkly Rep.2008;57:373–376.
- ,,, et al.Development of an electronic public health case report using HL7 v2.5 to meet public health needs.J Am Med Inform Assoc.2010;17:34–41.
Copyright © 2011 Society of Hospital Medicine